Alteryx Designer Desktop Discussions
Find answers, ask questions, and share expertise about Alteryx Designer Desktop and Intelligence Suite.- Community
- :
- Community
- :
- Participate
- :
- Discussions
- :
- Designer Desktop
- :
- Re: Variable importance plot - Boosted model - Val...
Variable importance plot - Boosted model - Values
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
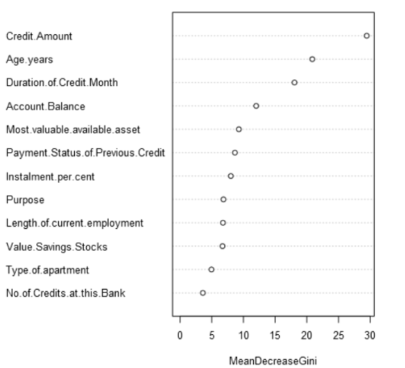
I am trying to know the values of Variables in the boosted Variable importance plot but it doesn't give that. How can I add values to Variable Importance plot so i can use them in further calculations?
Solved! Go to Solution.
- Labels:
-
Predictive Analysis
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @amrutas,
The image you've posted looks like it came from a Forest model. Are you trying to extract the variable importance values from a boosted model, random forest, or both? I ask because the process to do so will be slightly different depending on the model.
Either way, at a high level you will want to:
1. Connect an R tool to the O anchor of the tool you used to generate your model. The O stands for Object, and this is where the model object is streamed onto the canvas.
2. Write R code into the tool to read in the model and unserialize the object.
3. Load in the R package that corresponds to your model, this is randomForest for a random forest model and gbm for a boosted model.
4. Apply the corresponding R function to the model to extract the variable importance metric. This is importance() for a random forest and summary() for boosted models.
5. Write the resulting data back out to Alteryx.
The code to do this for a random forest model:
# Read in serialized model object
modelObj <- read.Alteryx("#1", mode="data.frame")
# Unserialize model
model <- unserializeObject(as.character(modelObj$Object[1]))
# load library
library(randomForest)
# extract feature importance
importance <- importance(model)
# convert to data frame
output <- data.frame(var = row.names(importance), importance)
# write out data frame
write.Alteryx(output, 1)
The code to do this for a boosted model:
# Read in serialized model object
modelObj <- read.Alteryx("#1", mode="data.frame")
# Unserialize model
model <- unserializeObject(as.character(modelObj$Object[1]))
# load package
library(gbm)
# extract variable importance
output <- summary(model)
# write out to Alteryx
write.Alteryx(output, 1)
I've attached a workflow that demonstrates the process for both a boosted model and a random forest model. Please be sure to read the documentation on each of the functions used to extract the variable importance metrics (hyperlinked above).
Hope this helps!
Sydney
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hey Sydney, your R code works to extract the values for the variable importance, thank you! My question is related to the other variable importance measure besides Mean Decrease Gini (IncNodePurity), which is Mean Decrease Accuracy (%IncMSE). Would you be able to construct an R code that derives those values for a Forest Model? I've read that the Gini index is biased towards values with numerous split points and wanted to use the %IncMSE to add another layer to my analysis.
*Not sure if you remember we, we met at the conference in Nashville and you helped me with the Importance Weights tool
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @h_kee,
Great to hear from you! Were you able to get the Importance Weights tool working on your machine?
Regarding your question, according to the documentation the same function (importance()) should be able to extract both mean decrease in accuracy and well as mean decrease in node impurity. Adding the argument type = 1 will extract mean decrease in accuracy, and type = 2 returns mean decrease in node impurity. Not specifying a type argument (as done in the example code in this thread) returns all importance metrics associated with the model.
The reason only the mean decrease in node impurity is getting returned by the code in this thread is because it is the only importance metric attached to the random forest model object coming out of the Forest Model tool.
I checked code that generates the random forest models in the Forest Model tool, and found that the mean decrease in accuracy metric isn't calculated because the argument importance = TRUE is not set. I modified the Forest Model tool so that the importance = TRUE argument is set, and the code in this thread works as expected for extracting mean decrease accuracy.
I've attached an example workflow with the modified Forest Model tool and working R code. You should be able to use the modified Forest Model tool included in the packaged workflow without any trouble. That being said - because I've changed the code in it, this tool is not officially supported (so don't overwrite your default Forest Model tool with it.)
Thanks!
Sydney
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
I was not able to get that tool to work but it's fine; you were very helpful though!
I have Alteryx 2019.1, which isn't the latest version, so I'm not able to download the modified Forest Model tool. Not to burden you with more work but if you're able to make it compatible with that version that'll be very helpful, if not, I can see about getting the latest version installed. Thanks again for your help, you are brilliant to say the least!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @h_kee,
I down-versioned the packaged workflow to 2018.4 - you should be able to open it without issue now, but please let me know if anything weird happens 🙂
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hey Sydney,
Your answer also helps me though I have another luxury question for general understanding.
Is it possible to work with Python coding module in order to do what you have written there or does it have to be a R customization module?
Best,
Atamert
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @atamertarslan,
All of the tools in the current predictive suite (the "brown tools" as well as the time series tools, AB testing tools, and prescriptive tools) are primarily written in the R programming language. The model object output of a predictive tool is a serialized R object, so if you'd like to work with the model objects generated by those tools, you will need to do so in R.
You can actually see the underlying R code for any of the predictive tools by right-clicking on the predictive tool and selecting the option Open Macro from the drop-down menu. This will open the workflow that executes the logic of the tool. The bulk of most predictive tool's functionality comes from an R tool within the macro.
Does this answer your question? Please let me know if there is anything I can expand on to help your understanding.
Thanks,
- Sydney
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi Sydney,
Is this possible for SVM? Is this a part of the output?
I haven't been abled to extract the values, but then again R isn't something I'm that familiar with...
Alexander.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @Alexandersd,
The variable importance metric is not a part of the SVM algorithm (it is a metric included by default for random forest and boosted models), but you can use the same general process to extract other values and metrics from the SVM tool. You can reach more about using custom R code to extract additional information from the predictive tools here.
If you are looking to determine variable importance for an SVM model, these resources also might be of interest to you:
https://stats.stackexchange.com/questions/2179/variable-importance-from-svm
Thanks,
Sydney
-
Academy
6 -
ADAPT
2 -
Adobe
204 -
Advent of Code
3 -
Alias Manager
78 -
Alteryx Copilot
25 -
Alteryx Designer
7 -
Alteryx Editions
95 -
Alteryx Practice
20 -
Amazon S3
149 -
AMP Engine
252 -
Announcement
1 -
API
1,208 -
App Builder
116 -
Apps
1,360 -
Assets | Wealth Management
1 -
Basic Creator
15 -
Batch Macro
1,559 -
Behavior Analysis
246 -
Best Practices
2,695 -
Bug
719 -
Bugs & Issues
1 -
Calgary
67 -
CASS
53 -
Chained App
268 -
Common Use Cases
3,825 -
Community
26 -
Computer Vision
86 -
Connectors
1,426 -
Conversation Starter
3 -
COVID-19
1 -
Custom Formula Function
1 -
Custom Tools
1,938 -
Data
1 -
Data Challenge
10 -
Data Investigation
3,487 -
Data Science
3 -
Database Connection
2,220 -
Datasets
5,222 -
Date Time
3,227 -
Demographic Analysis
186 -
Designer Cloud
741 -
Developer
4,372 -
Developer Tools
3,530 -
Documentation
527 -
Download
1,037 -
Dynamic Processing
2,939 -
Email
928 -
Engine
145 -
Enterprise (Edition)
1 -
Error Message
2,258 -
Events
198 -
Expression
1,868 -
Financial Services
1 -
Full Creator
2 -
Fun
2 -
Fuzzy Match
712 -
Gallery
666 -
GenAI Tools
3 -
General
2 -
Google Analytics
155 -
Help
4,708 -
In Database
966 -
Input
4,293 -
Installation
361 -
Interface Tools
1,901 -
Iterative Macro
1,094 -
Join
1,958 -
Licensing
252 -
Location Optimizer
60 -
Machine Learning
260 -
Macros
2,864 -
Marketo
12 -
Marketplace
23 -
MongoDB
82 -
Off-Topic
5 -
Optimization
751 -
Output
5,255 -
Parse
2,327 -
Power BI
228 -
Predictive Analysis
937 -
Preparation
5,169 -
Prescriptive Analytics
206 -
Professional (Edition)
4 -
Publish
257 -
Python
855 -
Qlik
39 -
Question
1 -
Questions
2 -
R Tool
476 -
Regex
2,339 -
Reporting
2,433 -
Resource
1 -
Run Command
575 -
Salesforce
277 -
Scheduler
411 -
Search Feedback
3 -
Server
630 -
Settings
935 -
Setup & Configuration
3 -
Sharepoint
627 -
Spatial Analysis
599 -
Starter (Edition)
1 -
Tableau
512 -
Tax & Audit
1 -
Text Mining
468 -
Thursday Thought
4 -
Time Series
431 -
Tips and Tricks
4,187 -
Topic of Interest
1,126 -
Transformation
3,730 -
Twitter
23 -
Udacity
84 -
Updates
1 -
Viewer
3 -
Workflow
9,980
- « Previous
- Next »