Alteryx Designer Desktop Discussions
Find answers, ask questions, and share expertise about Alteryx Designer Desktop and Intelligence Suite.- Community
- :
- Community
- :
- Participate
- :
- Discussions
- :
- Designer Desktop
- :
- .xyz file input
.xyz file input
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Does anyone have any experience using scientific .xyz files? They're used in sciences (something like computational chemistry to describe a molecular structure), but apparently there's not really a standard format, and you can incorporate comments throughout, I'm not really sure how to parse it consistently.
I attached one as a .csv, but I've got millions of these, and wasn't sure if I can just parse it like a general .csv, or if I have to get metadata from where the data is coming from to determine the parsing.
Bottom line, if anyone with some sort of a science background can point me in the right direction of a good resource on working with these, I can't really find much on this subject. I'm decent with R and Python as well, so if you have any insight on packages there, that would be useful.
Solved! Go to Solution.
- Labels:
-
Parse
-
Preparation
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Could you post some example files with varying style and the desired output? The core structure seems simple enough that Alteryx should be more than capable of parsing variations on that core (The "general .csv" parsing approach you mentioned).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
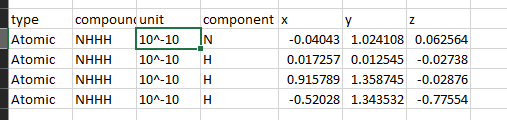
@CharlieS Thanks for the response, I copied and pasted some of the .xyz files - the image is what I'd be looking for; it's more involved than text parsing based on spaces, I'm trying to figure out things such as
1) how do I determine units, does that come from metadata? I'm not confident they're all angstrom (10^-10)
2) Is the scientific software generating this by just pulling from a big library of stock metadata, or calculating these independently
Basically I've built a bunch of prediction tools and apps within Alteryx for myself, so it would be helpful to use these .xyz files within Alteryx (as opposed to the incredibly clunky lab software), as the end result is a simple 3-d axis (just tons of them), but I can't find many resources on how I associate them with metadata and trusting that they're consistent and parsed correctly. I might be biting off more than I can chew right now though, but if someone with a computational science background can point me to a good resource, that would be helpful.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
So here's something I put together to get things started. I used the file name to determine how the file needed to be parsed (looking for "_fcs" or "_acs"). After that, I parsed the necessary information from each file (compound name, component, bond lengths). I figured the positions/bond lengths could be determined by values, so this definitely needs a chemists' insight. In the attached wizard/app below, there's a collapsed tool container with this work.
When I was done with that, it was obvious that a lot of redundancies could be consolidated, so I did that and wrapped it up into a wizard/app. This wizard allows you to input the file path and it will write a .yxdb of the requested information in the same folder.
I'm sure there's a million things that need to be added before this is ready, but it should give you some good ideas to get started.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Thanks a lot for taking the time to put that together! I'm going through it right now and tweaking some things and incorporating it with some other sections that I'm working on. I marked it as a solution, I'm hoping over the long-term that I can build an entire science application within alteryx, that works much better than scientific software from the late 90's.
-
Academy
6 -
ADAPT
2 -
Adobe
204 -
Advent of Code
3 -
Alias Manager
78 -
Alteryx Copilot
26 -
Alteryx Designer
7 -
Alteryx Editions
95 -
Alteryx Practice
20 -
Amazon S3
149 -
AMP Engine
252 -
Announcement
1 -
API
1,208 -
App Builder
116 -
Apps
1,360 -
Assets | Wealth Management
1 -
Basic Creator
15 -
Batch Macro
1,559 -
Behavior Analysis
246 -
Best Practices
2,695 -
Bug
719 -
Bugs & Issues
1 -
Calgary
67 -
CASS
53 -
Chained App
268 -
Common Use Cases
3,825 -
Community
26 -
Computer Vision
86 -
Connectors
1,426 -
Conversation Starter
3 -
COVID-19
1 -
Custom Formula Function
1 -
Custom Tools
1,938 -
Data
1 -
Data Challenge
10 -
Data Investigation
3,487 -
Data Science
3 -
Database Connection
2,220 -
Datasets
5,222 -
Date Time
3,227 -
Demographic Analysis
186 -
Designer Cloud
742 -
Developer
4,372 -
Developer Tools
3,530 -
Documentation
527 -
Download
1,037 -
Dynamic Processing
2,939 -
Email
928 -
Engine
145 -
Enterprise (Edition)
1 -
Error Message
2,258 -
Events
198 -
Expression
1,868 -
Financial Services
1 -
Full Creator
2 -
Fun
2 -
Fuzzy Match
712 -
Gallery
666 -
GenAI Tools
3 -
General
2 -
Google Analytics
155 -
Help
4,708 -
In Database
966 -
Input
4,293 -
Installation
361 -
Interface Tools
1,901 -
Iterative Macro
1,094 -
Join
1,958 -
Licensing
252 -
Location Optimizer
60 -
Machine Learning
260 -
Macros
2,864 -
Marketo
12 -
Marketplace
23 -
MongoDB
82 -
Off-Topic
5 -
Optimization
751 -
Output
5,255 -
Parse
2,328 -
Power BI
228 -
Predictive Analysis
937 -
Preparation
5,169 -
Prescriptive Analytics
206 -
Professional (Edition)
4 -
Publish
257 -
Python
855 -
Qlik
39 -
Question
1 -
Questions
2 -
R Tool
476 -
Regex
2,339 -
Reporting
2,434 -
Resource
1 -
Run Command
575 -
Salesforce
277 -
Scheduler
411 -
Search Feedback
3 -
Server
630 -
Settings
935 -
Setup & Configuration
3 -
Sharepoint
627 -
Spatial Analysis
599 -
Starter (Edition)
1 -
Tableau
512 -
Tax & Audit
1 -
Text Mining
468 -
Thursday Thought
4 -
Time Series
431 -
Tips and Tricks
4,187 -
Topic of Interest
1,126 -
Transformation
3,730 -
Twitter
23 -
Udacity
84 -
Updates
1 -
Viewer
3 -
Workflow
9,980
- « Previous
- Next »