Tool Mastery
Explore a diverse compilation of articles that take an in-depth look at Designer tools.- Community
- :
- Community
- :
- Learn
- :
- Academy
- :
- Tool Mastery
- :
- Tool Mastery | Decision Tree
Tool Mastery | Decision Tree
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Notify Moderator
03-15-2018 01:36 PM - edited 08-03-2021 03:44 PM

This article is part of the Tool Mastery Series, a compilation of Knowledge Base contributions to introduce diverse working examples for Designer Tools. Here we’ll delve into uses of the Decision Tree Tool on our way to mastering the Alteryx Designer:
The subtitle to this article should be a short novel on configuring the Decision Tree Tool in Alteryx. The initial configuration of the tool is very simple, but it you chose to customize the configuration of the tool at all, it can get complicated quickly. In this article, I am focusing on the configuration of the Tool. However, because it is a Tool Mastery, I am covering everything within the configuration of the tool.
If you're looking for something other than a detailed overview of configuring the tool, there are a few other resources on Community for you to check out!For a really amazing overview of using the tool, check out the Data Science Blog article; An Alteryx Newbie Works Through the Predictive Suite: Decision Tree. If you're looking for a more general introduction on Decision Trees, read Planting Seeds – an Introduction to Decision Trees, and if you’re interested more in the outputs of the Decision Tree Tool, please take a look at Understandingthe Outputs of the Decision Tree Tool.
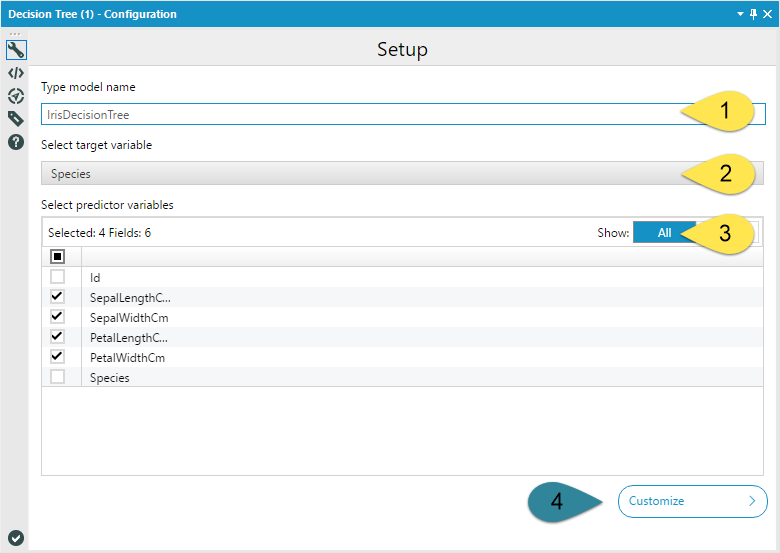
If you're still with me, lets get going! Like I mentioned, the basic configuration of the Decision Tree Tool is straight forward. The Model Name (1) is a text input. Any spaces you include in your model name will be replaced by underscores so that R can handle the name. The target variable (2) is the variable you are predicting, and the predictor variables (3) are the fields you are going to use to predict your target variable. You should select one or more fields.
Do not include unique identifier fields. They have no predictive value and can cause runtime errors. It is also important at this point to make sure all of your model variables have appropriate data types. Make sure that your continuous variables are not string, and that your categorical variables represented by numbers are string and not numeric.
If you so choose, you can leave the Decision Tree Tool configuration at that. If you choose to open model customization (4), the tool gets a little more complicated…
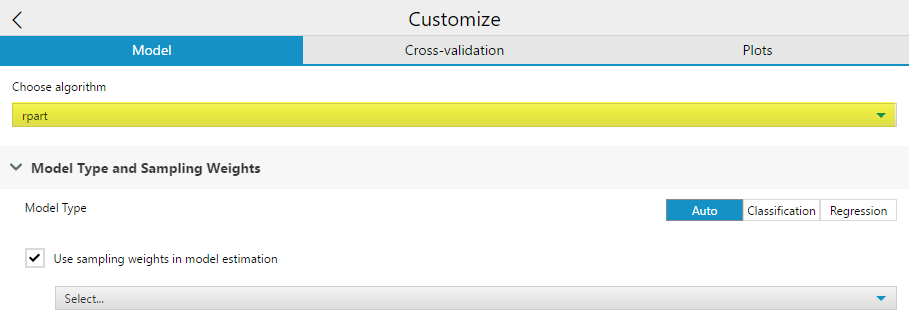
First, you have the option to select which rpackage (and underlying algorithm) your decision tree is created with, rpartor c5.0. Each of these two packages correspond to a different algorithm.If you are using the Decision Tree Tool for In-DB processing on a SQL Server, a third rpackage, RevoScaleRis used to construct the decision tree.
The rpart (recursive partitioning) package implements the functionality from the book Classification and Regression Treesby Breiman, Freidman, Olshen and Stone, published in 1984. A thorough introduction to rpart can be found here.
The C5.0 R package fits Quinlan’s C5.0 classification model based on the source www.rulequest.com.An informal tutorial on C5.0 from rulequest is available here.
The C5.0 algorithm will not construct regression trees (continuous target variables). rpart will construct both classification and regression trees.
In the Decision Tree Tool, the options in Customize Model will change based on which algorithm you select.
rpart
If you choose the rpart algorithm, your customization drop-down options are Model Type and Sampling Weights, Splitting Criteria and Surrogates, and HyperParameters
Model Type allows you to specify either Auto, Classification, or Regression, which will determine what type of decision tree you construct. Auto will select either Classification or Regression based on your target variable data type.
Checking the Use Sampling weights in model estimation option will allow you to specify an importance weight field for the decision tree to account for. If you use a field both as a predictor and a sample weight, the output weight variable field will have “Right_” appended to the field name as a prefix.
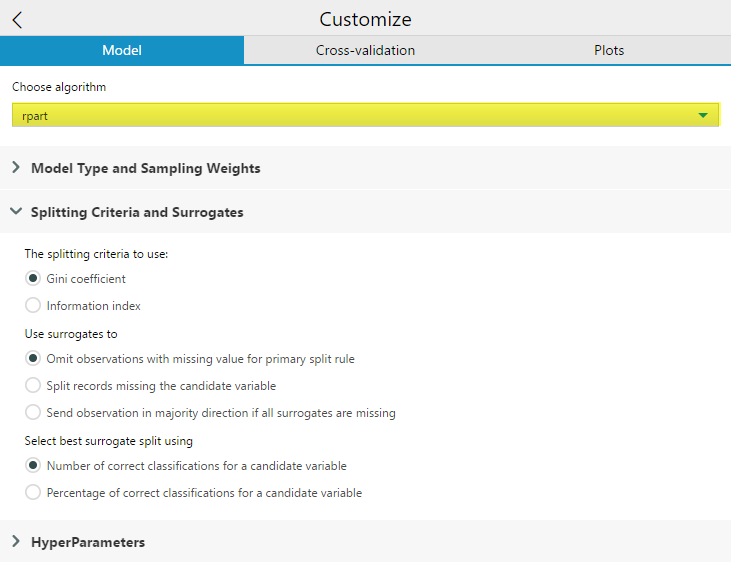
Splitting Criteria to use will appear if you are building a classification tree (and have selected Model Type: Classification in the Model Type and Sampling Weights Drop Down). If you’re building a classification tree, but the model type is set to Auto, the option will not be available. If you are running a regression model the splitting criterion will automatically be Least Squares. For this option, you can select which splitting criteria to use, Gini impurity or Information Index.
The Use surrogates to option allows you to select a method for using surrogates in the splitting process. In Decision Trees,a surrogate is a substitute predictor variable and threshold that behaves similarly to the primary variable and can be used when the primary splitter of a node has missing data values. Surrogates can also be used to reveal common patterns among predictors variables in the data set. Omit observations with missing value for primary split rule will exclude any records with missing values for the variable being used to split that node from being passed further down into the tree. Split records missing the candidate variable will split records missing a value for a variable used at a node with the surrogate. If all surrogates are missing, then the observation will not be passed down. Send observation in majority direction is all surrogates are missing handles the condition where all surrogates are missing by sorting the record in the direction of the majority of records being split.
The Select best surrogate split using allows you to choose the criteria the rpart algorithm uses to select a variable to act as a surrogate. The default is the total Number of correct classifications for a candidate variable. Percentage of correct classifications for a candidate variable is calculated over the non-missing values of the surrogate. Number of correct classifications for a candidate variable more severely penalizes predictor variables with a large number of missing values.
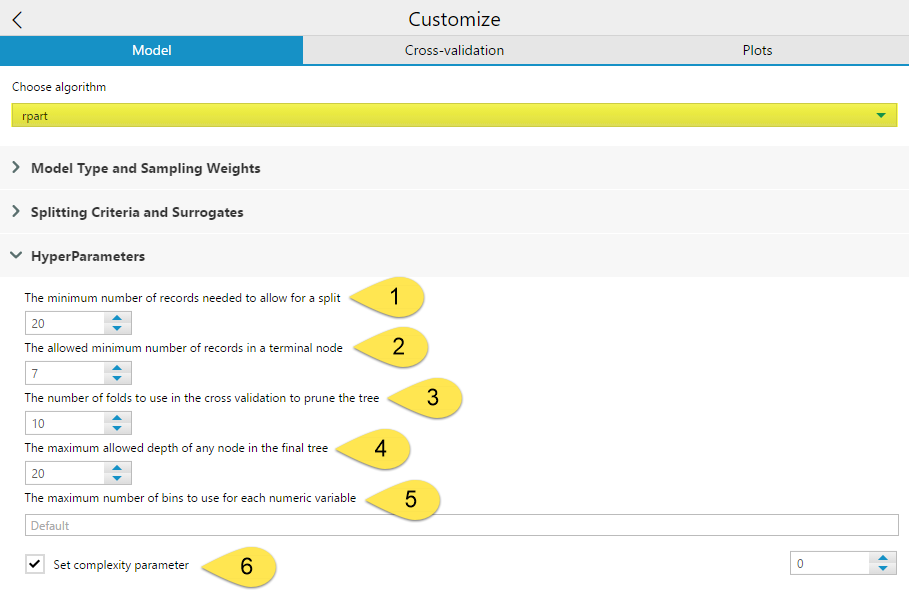
The HyperParameters include six different settings you can configure.
The minimum number of records need to allow for a split (1) allows you to specify the minimum number of records in a node needed for a split to occur. The allowed minimum number of records in a terminal node (2) is where you can specify the minimum number of records needed for a terminal node to exist. Having lower numbers set for (1) and (2) increases the potential number of final terminal nodes. Conversely, having too high a value set for the (2) parameter will cause premature terminal nodes that could potentially further subdivided for accuracy. By default, the (2) value is one-third of the value specified for (1). Setting the (1) too high can also potentially prevent maximum accuracy or cause your model to fail. However, having the value set too low will cause over-fitting of your training data. You want small enough groups that you are getting accurate results, while still being broad enough that the splitting rules will work on other data sets. As a rule, it is not advisable to attempt to split a node with fewer than 10 records in it.
The number of folds to use in the cross validation to prune the tree (3) allows you to select the number of groups the data should be divided into for testing the model. By default this number is 10, but other common values are 5 and 20. A higher number improves accuracy but also increases run time. If a tree is pruned using a complexity parameter, cross-validation determines how many branches are in a tree. In cross validation, N - 1 of the folds are used to create a model, and the other fold is used as a sample to determine the number of branches that best fits the holdout fold in order to avoid overfitting.
The maximum allowed depth of any node in the final tree (4) sets the maximum number of levels of branches allowed from the root node to the most distance node from the root to limit the overall size of the tree. The root is counted as depth 0.
The maximum number of bins to use for each numeric variable (5) option is only relevant when using the decision tree tool on a SQL Server in-DB data stream. In this case, the RevoScaleR function from the rxDTree rpackage is used to create a scalable decision tree, which can handle numeric variables using an equal interval binning process to reduce the computation complexity. This value sets the number of bins to use for each variable. The default value uses a formula based on the minimum number of records needed to allow for a split.
The Set complexity parameter (6) option is the value that controls the size of the decision tree, by preventing any split that does not improve the model by at least a factor of the complexity parameter (cp) from being constructed. A smaller value results in more branches in the tree, and a larger value results in less branches. If a complexity parameter is not selected, it is automatically determined based on cross-validation.
C5.0
If you selected the C5.0 algorithm, your customization drop-down options are Structural Options, Detailed Options, and Numerical Hyperparameters.
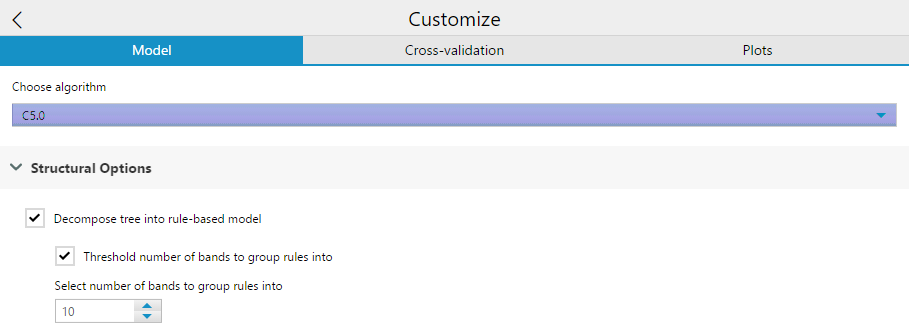
Structural options include a check-box to Decompose tree into rule-based model. Essentially, this option changes the structure of the output algorithm from a decision tree (the default) into a collection of rules. It also grants the ability to generate classifiers called rulesets that consist of unordered collections of (relatively) simple if-then rules.
Selecting this creates a second check-box option to Threshold the number of bands the group rules into. If you select this option, you then need to specify the number of bands to group the rules into. This is a number used to simplify the model.
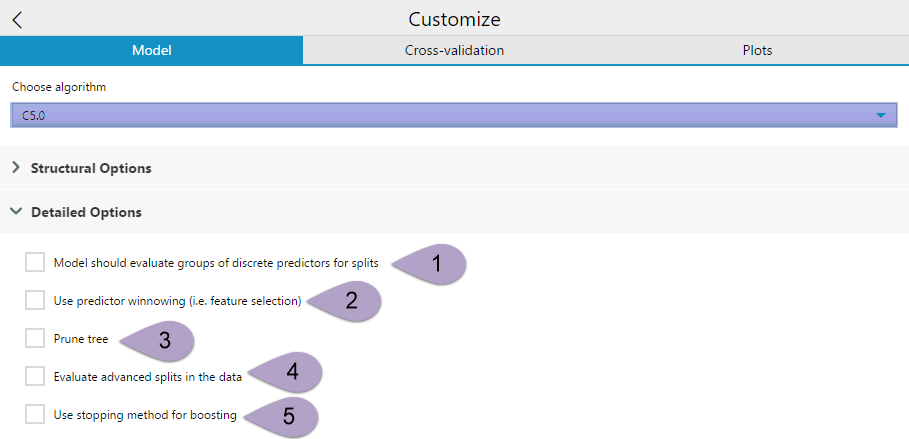
The Detailed Options drop-down has five check-box options.
The option Model should evaluate groups of discrete predictors for splits (1) can be selected to simplify the model and reduce overfitting by grouping categorical predictor variables together. This option is recommended when there are important discrete attributes that have more than four or five values.
Use predictor winnowing(2) performs feature selection on your predictor variables, separating the useful variables from the less-useful variables, thereby simplifying your model. Winnowing pre-selects a subset of the predictor variables that will be used to construct the decision tree or ruleset.
The Prune tree (3) option can be selected to enable global pruning for the decision tree model. Pruning simplifies the tree and can reduce overfitting. There are situations where it is beneficial to leave global pruning turned off, particularly when you are generating rulesets instead of a tree.
Evaluate advanced splits in the data (4) is an option in C5.0 that allows the model to evaluate possible advanced splits in the data. Advanced splitsare also referred to as Fuzzy Thresholds or Soft Thresholds. In the C5.0 algorithm, each split threshold in the decision tree actually consists of three parts -- a lower bound (lb), an upper bound (ub), and an intermediate value (t). The intermediate value is what is displayed as the splitting threshold in the tree.
If you enable the Evaluate advanced splits in the data option, each threshold in the tree changes to the form = ub (t). When variable value is outside of the lower (lb) and upper bounds (ub), the record is sorted using the single branch corresponding to the `' result, but if the value is between the lower and upper bound, the record is passed to both branches of the tree, and the result is combined. The advanced splits option only effects the decision trees, it will not change your model if you have chosen to create a ruleset.
The Use stopping method for boosting (5) enables the model to evaluate if boosting iterations are becoming ineffective and, if so, stops boosting.
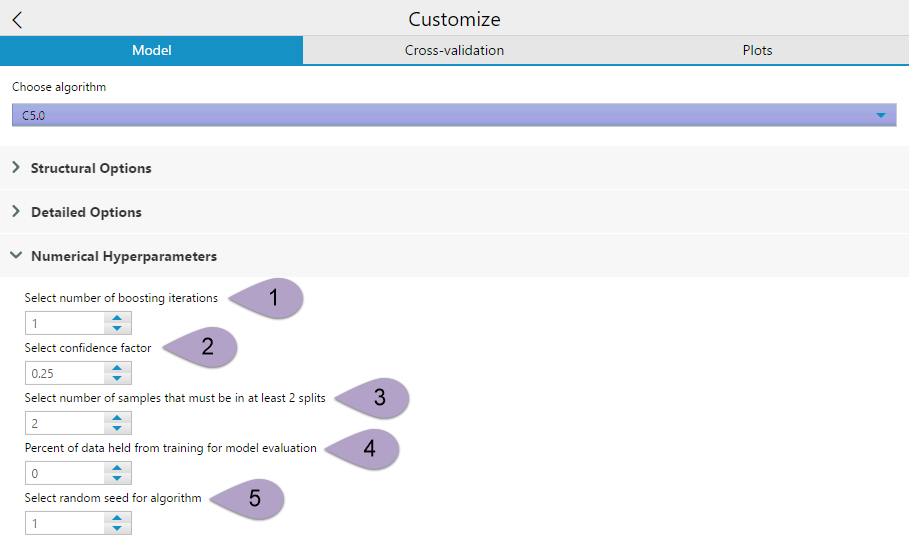
Numerical Hyperparameters also has five options for you to specify from.
Select number ofboostingiterations (1). This option allows you to determine the number of boosting iterations run to generate your decision tree. Selecting 1 creates a single model. This will continue for whatever number you have specified. The boosting iterations stop if the latest tree is extremely accurate or inaccurate. More computation with a greater number of iterations, but 10-classifier boosting reduces error for test cases an average of 25%.
Select confidence factor (2) can be a value between 0 and 1. This setting affects the way that the error rates are estimated, thereby impacting the level of pruning performed. Values less than the default, 0.25 cause more of the initial tree to be pruned, while larger values equate to less pruning. This setting is analogous to rpart’s complexity parameter.
Select number of samples that must be in at least 2 splits (3) limits the level to which the initial tree can fit the data. At each branch point in the constructed tree this value must follow at least two of the daughter branches. A larger number gives a smaller, more simplified, tree.
Percent of data held from training for model evaluation (4) Select the portion of the data used to train the model. Use the default value 0 to use all of the data to train the model. Select a larger value to hold that percent of data from training and evaluation of model accuracy.
Select random seed for algorithm (5) allows you to make the randomized components of the decision tree model reproducible. It doesn’t matter what you set your seed to, as long as you keep it consistent.
And Now, the Other Two Tabs
The Cross-validation tab allows you to create estimates of model quality using cross-validation. If you would like to know more about configuring cross-validation, please take a look at the Community ArticleGuide to Cross-Validation.
Plots allows you to select the option of Displaying a static report, as well as adding a Tree Plot and, if you’re using the rpart algorithim a Prune Plot. By default, the static report option is checked. If you uncheck the Display static report option, there is no R (Report) output for your tool.
If you turn on the Tree Plot or Prune Plot options, each selected plot is appended to your R (Report) output. For the C5.0 algorithm, the Tree Plot will be appended to the end of the report, and you will need to click the next button at the top of the report window to find it.
There are many ways to configure the Decision Tree Tool, and I hope that this article has made each of these options a little clearer, and that you now feel ready to take on the Decision Tree Tool with confidence! For a review and explanation of the results of the Decision Tree Tool, check out this supporting article, Understanding the Decision Tree Tool Outputs.
By now, you should have expert-level proficiency with the Decision Tree Tool! If you can think of a use case we left out, feel free to use the comments section below! Consider yourself a Tool Master already? Let us know atcommunity@alteryx.comif you’d like your creative tool uses to be featured in the Tool Mastery Series.
Stay tuned with our latest posts every#ToolTuesdayby following@alteryxon Twitter! If you want to master all the Designer tools, considersubscribingfor email notifications.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Thanks for your post. I have a few questions.
1. What exactly is the difference between "Number of folds to use in cross validation to prune the tree" under the model tab and "Number of cross validation folds" under the cross-validation tab in decision tree tool?
2. Suppose I split my data into train and test set. Then I use the decision tree tool on my training set (with cross-validation) and finds out that level 8 with nsplits = 52 is the most optimal tree (based on min x errors). I have the impression that we move on from cross validation and proceed to use train the model with my newly found hyper parameters on the entire training data. How do I then move on from cross validation to train my model with these hyper parameters that I found out? In my new hyper parameter setting, do I set maxdepth = 8 and is that it?
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Hi @data2,
1. The Number of folds to use in cross-validation to prune the tree option under the model tab is related to a procedure for pruning the decision tree trained by the tool, and the Number of cross-validation folds option under the cross-validation tab is used for performing a cross-validation routine to evaluate the decision tree model. You might notice when the cross-validation option under the cross-validation tab is enabled, the "R" (report) output of the Decision Tree tool includes some additional plots - these are the additional evaluation metrics generated by performing cross-validation to evaluate the model.
So to answer your question, the first option determines how many sub-groups the training data is split up into for pruning, and the second option found under the cross-validation tab defines how many sub-groups the training data is split up into to help create estimates for evaluating the model. If you choose to perform cross-validation for evaluation (enabling cross-validation options in the cross-validation tab) a second cross-validation routine will be performed for each model that is trained to help prune the trained decision trees.
For help understanding how cross-validation is used to prune decision trees, you might find the following resources helpful:
To prune or not to prune: cross-validation is the answer
Slides from a Stats Lecture on Decision Trees
An Introduction to Recursive Partitioning Using the RPTART Routines - Part 4.2
Essentially, cross-validation is leveraged to determine which branches in a decision tree do not generalize to "new" data well and removes those branches to mitigate the risk of overfitting.
2. Using cross-validation to evaluate your model can eliminate the need to create separate training and test data sets, as they effectively serve the same purpose of providing a way to test your model against data it hasn't been exposed to during the training process. For hyperparameter tuning, you could just use the metrics generated by the cross-validation routine (performed using the options set under the cross-validation tabs) to determine optimal values and use the model trained with those values - the tool will use the entire dataset to train the final model. You can read more about cross-validation and hold-out datasets here.
I hope this helps!
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Hi @SydneyF, your explanation sure helped! Thanks for the detailed explanation.
Regarding to question 2, the focus of my question was really how do we then "use the model trained with those values" after finding out the optimal values of hyper parameter using cross validation?
From the decision tree (with cross validation enabled) output, say i know the optimal values are level 8, nsplit = 52 and complexity parameter = 0.003 (based on min x errors). How do i input all these parameters into the tuning of hyper parameters tab in decision tree tool for the final model?
From what I see, only setting max depth and complexity parameter are relevant? If so, then in this case
1. should i set max depth as 8
2. set cp as 0.003 (which is invalid on Alteryx since min increment must be 0.1)
Thank you!
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Hi @data2,
You shouldn't need to reconfigure your tool or retrain a model once you have cross-validation metrics you are satisfied with. The decision tree tool optimizes the structure of the models it generates based on the hyperparameters you set at the beginning and the training data. So, in your example, whatever you set from the start caused the model to land on a structure where the depth was 8 and the cp was 0.003.
Does this make sense? The model evaluation cross-validation isn't testing different hyperparameter values, it is evaluating a final model, given the hyperparameters you had already set in the tool. To test different hyperparameters in Alteyx, you will need to run many iterations of models using the Decision tree tool, and land on the "best" model based on the evaluation metrics. Whichever iteration of your tests results in the best metrics will already include a trained model, ready to use in the "O" anchor of the tool.
- Sydney
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Hi @SydneyF,
Yes that makes sense to me. What you are saying is that based on the hyperparameters I had configured right at the start, the model (run with cross validation) gives me a single optimal tree with values within the range of hyperparameters specified right?
1. What I am also curious would be how do we then test the different hyperparameters in Alteryx apart from guess and check? I understand that in python we can use Scikit Learn and tap on techniques like Grid Search/ Random Search etc. I also refer to your great post over here: https://community.alteryx.com/t5/Data-Science-Blog/Hyperparameter-Tuning-Black-Magic/ba-p/449289
In Alteryx, do we have a tool that allows us to do Grid Search or Random Search? Tried looking for these tools in the Gallery but can't seem to find any.
2. When tuning hyperparameters, shouldn't we still use cross validation (for example k-folds)? If we had chose to only hold out 1 validation and 1 test set, the data in test set might not be representative of the entire data set (can be extreme cases) then the model with tuned hyperparameters would not do well on the test set? Hence, it seems to me that using cross validation (k-folds) is a good idea to overcome this issue. And if you agree with me on this, can you help me understand if this cross validation is in fact different from the cross validation tab in the decision tree tool since you mentioned the model evaluation cross validation is not for testing different hyperparameters values.
Thank you!
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Hi @data2,
Currently, there is not any functionality in Alteryx to perform hyperparameter tuning beyond guess and check. If this is something you would like to see added to the product, please post an idea in our Ideas forum - please let me know if you do and I will be sure to star it!
To answer your second question, yes it is reasonable and advisable to use cross-validation to assess different hyperparameter values. The cross-validation tab in the Decision Tree tool can be used for this purpose. The cross-validation routine is used to evaluate the performance of a model, so to leverage it to test different hyperparameter values you would:
1. Set up multiple Decision Tree tools with different hyperparameter values configured in the tool's advanced settings. This is effectively the "guess and check" method for hyperparameter tuning
2. Enable the cross-validation evaluation routine in the cross-validation tab.
3. Run the model(s)
4. Compare the cross-validation metrics to determine which hyperparameter values resulted in the most optimal model. Use that model and rejoice.
When I said that the cross-validation routine is not testing different hyperparameters values, I meant just that. It can be used to evaluate the efficacy of different hyperparameter values, but the cross-validation routine is not testing different hyperparameters values for you - you need to do that manually by configuring the tool to use different hyperparameter values.
Another option would be to use the Cross-Validation tool available on the Alteryx Gallery instead of using the cross-validation routine embedded in the Decision Tree tool. The benefit of using this tool is that you could have the metrics returned side-by-side in a single report as opposed to multiple reports.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Hi @SydneyF
Thank you for your answers! You have been very helpful. You have cleared many of my doubts in our conversation with your fast replies. I have also posted an idea titled "Grid Search/ Random Search CV". Would be thrilled if Alteryx provides such a tool one day.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Thanks - this helped a lot.
One question - I am running an analysis of daily continuous data for which I am trying to compare different time series and classification models.
As such, when I run the tool (and also for the Random Forest and Boosted Model), what interval are the results for? I.e. is the model trying to predict the value for the next period or p+? And, is there a setting where I can change this?
Thanks
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Notify Moderator
Great article and follow-up. Thank you.
-
2018.3
1 -
2023.1
1 -
API
2 -
Apps
7 -
AWS
1 -
Configuration
4 -
Connector
4 -
Container
1 -
Data Investigation
11 -
Database Connection
2 -
Date Time
4 -
Designer
2 -
Desktop Automation
1 -
Developer
8 -
Documentation
3 -
Dynamic Processing
10 -
Error
4 -
Expression
6 -
FTP
1 -
Fuzzy Match
1 -
In-DB
1 -
Input
6 -
Interface
7 -
Join
7 -
Licensing
2 -
Macros
7 -
Output
2 -
Parse
3 -
Predictive
16 -
Preparation
16 -
Prescriptive
1 -
Python
1 -
R
2 -
Regex
1 -
Reporting
12 -
Run Command
1 -
Spatial
6 -
Tips + Tricks
1 -
Tool Mastery
99 -
Transformation
6 -
Visualytics
1
- « Previous
- Next »