Alteryx Designer Desktop Discussions
Find answers, ask questions, and share expertise about Alteryx Designer Desktop and Intelligence Suite.- Community
- :
- Community
- :
- Participate
- :
- Discussions
- :
- Designer Desktop
- :
- Encoding Error in R tool
Encoding Error in R tool
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
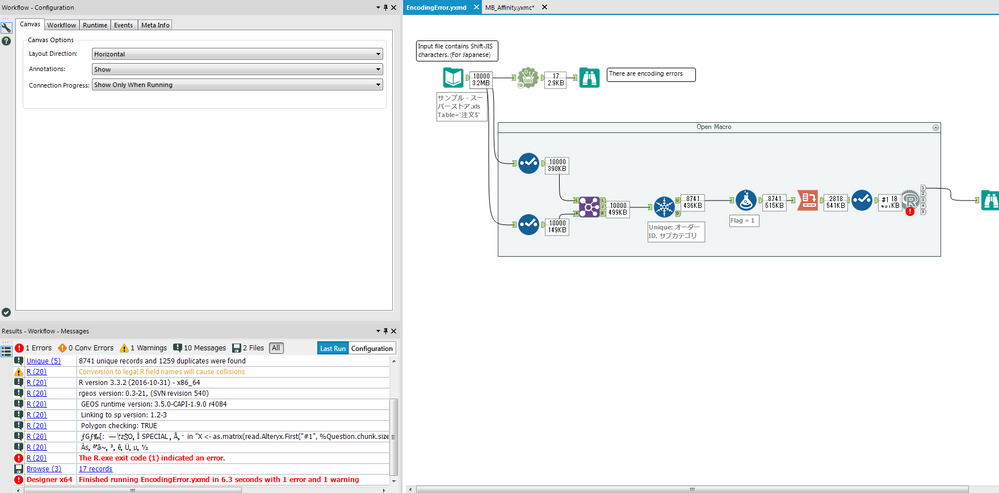
I want to use R-tools for files in Japanese, encode is Shift-JIS.
I think I have to use encoding='cp932' parameter in R script.
But it doesn't work by my trials...
I attached my flow.
Would someone please check my workflow?
Thanks,
Toshi
Solved! Go to Solution.
- Labels:
-
R Tool
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
@SydneyF - I have an R-script that I am using to upload data to a database, however I am running into issues with certain header names that have special characters. I tried some of the work arounds in this chain with no success. The base code is pasted below and the errors are when a header field has /, -, etc. All special characters (inclusing spaces) seem to be changed to . when pulled into the R tool. How can I get them to retain the special characters for the headers?
Thanks,
Matt
if("RPostgreSQL" %in% rownames(installed.packages(),) == FALSE)
{install.packages("RPostgreSQL",repos="http://cran.revolutionanalytics.com")}
library (RPostgreSQL)
value <- read.Alteryx("#4", mode="data.frame")
names(value) <- gsub(x = names(value), "[.]", " ", value)
drv <- dbDriver("PostgreSQL")
con <- dbConnect(drv,
dbname = "XXXXXXX",
host = "XXX.XX.XX.XX",
port = XXXX,
user = "XXXXXXX",
password = "XXXXXXX")
RPostgreSQL::dbSendQuery(con, "TRUNCATE schema.tablename")
name <- c("schema", "tablename")
dbWriteTable(
con = con,
name = name,
value = value,
row.names = FALSE,
append = TRUE,
col.names = FALSE
)
dbDisconnect(con)
write.Alteryx(value, 1)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @MatthewYoung,
When you read data into the R Tool, it creates an R data frame. The column names in an R data frame must be syntactically valid, which for R means that they must start with a letter, and include only letters, numbers, and the special characters . or _. Forward slashes, dashes, spaces, and any other special characters that are not syntactically valid in R will all be converted to a dot.
There are some ways to work around this when using other functions, like read.csv() or data.frame(), however, there is not currently an argument in read.Alteryx() that can be applied to retain the special characters. It looks like someone has posted a product idea for enhancements to the read.Alteryx() function here - I would suggest starring it and commenting with your use case if you would like to see updates to this function as well. Our product managers are very active on the Product Ideas page and are always looking for great new features to add to our software. The more attention an idea gets (i.e., stars and comments) the more likely it is to be considered.
If you would like to read more about special characters in R, please see the following Stackoverflow threads:
https://stackoverflow.com/questions/46437258/why-r-changes-special-characters-in-column-name-to-dots
https://stackoverflow.com/questions/42842031/r-trouble-with-space-and-in-variable-and-column-name
Thanks!
SydneyF
- « Previous
- Next »
-
AAH
1 -
AAH Welcome
2 -
Academy
24 -
ADAPT
82 -
Add column
1 -
Administration
20 -
Adobe
176 -
Advanced Analytics
1 -
Advent of Code
5 -
Alias Manager
70 -
Alteryx
1 -
Alteryx 2020.1
3 -
Alteryx Academy
3 -
Alteryx Analytics
1 -
Alteryx Analytics Hub
2 -
Alteryx Community Introduction - MSA student at CSUF
1 -
Alteryx Connect
1 -
Alteryx Designer
44 -
Alteryx Engine
1 -
Alteryx Gallery
1 -
Alteryx Hub
1 -
alteryx open source
1 -
Alteryx Post response
1 -
Alteryx Practice
134 -
Alteryx team
1 -
Alteryx Tools
1 -
AlteryxForGood
1 -
Amazon s3
138 -
AMP Engine
191 -
ANALYSTE INNOVATEUR
1 -
Analytic App Support
1 -
Analytic Apps
17 -
Analytic Apps ACT
1 -
Analytics
2 -
Analyzer
17 -
Announcement
4 -
API
1,040 -
App
1 -
App Builder
43 -
Append Fields
1 -
Apps
1,168 -
Archiving process
1 -
ARIMA
1 -
Assigning metadata to CSV
1 -
Authentication
4 -
Automatic Update
1 -
Automating
3 -
Banking
1 -
Base64Encoding
1 -
Basic Table Reporting
1 -
Batch Macro
1,271 -
Beginner
1 -
Behavior Analysis
217 -
Best Practices
2,414 -
BI + Analytics + Data Science
1 -
Book Worm
2 -
Bug
622 -
Bugs & Issues
2 -
Calgary
59 -
CASS
46 -
Cat Person
1 -
Category Documentation
1 -
Category Input Output
2 -
Certification
4 -
Chained App
235 -
Challenge
7 -
Charting
1 -
Clients
3 -
Clustering
1 -
Common Use Cases
3,389 -
Communications
1 -
Community
188 -
Computer Vision
45 -
Concatenate
1 -
Conditional Column
1 -
Conditional statement
1 -
CONNECT AND SOLVE
1 -
Connecting
6 -
Connectors
1,180 -
Content Management
8 -
Contest
6 -
Conversation Starter
17 -
copy
1 -
COVID-19
4 -
Create a new spreadsheet by using exising data set
1 -
Credential Management
3 -
Curious*Little
1 -
Custom Formula Function
1 -
Custom Tools
1,722 -
Dash Board Creation
1 -
Data Analyse
1 -
Data Analysis
2 -
Data Analytics
1 -
Data Challenge
83 -
Data Cleansing
4 -
Data Connection
1 -
Data Investigation
3,062 -
Data Load
1 -
Data Science
38 -
Database Connection
1,898 -
Database Connections
5 -
Datasets
4,578 -
Date
3 -
Date and Time
3 -
date format
2 -
Date selection
2 -
Date Time
2,880 -
Dateformat
1 -
dates
1 -
datetimeparse
2 -
Defect
2 -
Demographic Analysis
173 -
Designer
1 -
Designer Cloud
473 -
Designer Integration
60 -
Developer
3,646 -
Developer Tools
2,919 -
Discussion
2 -
Documentation
454 -
Dog Person
4 -
Download
907 -
Duplicates rows
1 -
Duplicating rows
1 -
Dynamic
1 -
Dynamic Input
1 -
Dynamic Name
1 -
Dynamic Processing
2,539 -
dynamic replace
1 -
dynamically create tables for input files
1 -
Dynamically select column from excel
1 -
Email
743 -
Email Notification
1 -
Email Tool
2 -
Embed
1 -
embedded
1 -
Engine
129 -
Enhancement
3 -
Enhancements
2 -
Error Message
1,977 -
Error Messages
6 -
ETS
1 -
Events
178 -
Excel
1 -
Excel dynamically merge
1 -
Excel Macro
1 -
Excel Users
1 -
Explorer
2 -
Expression
1,696 -
extract data
1 -
Feature Request
1 -
Filter
1 -
filter join
1 -
Financial Services
1 -
Foodie
2 -
Formula
2 -
formula or filter
1 -
Formula Tool
4 -
Formulas
2 -
Fun
4 -
Fuzzy Match
614 -
Fuzzy Matching
1 -
Gallery
590 -
General
93 -
General Suggestion
1 -
Generate Row and Multi-Row Formulas
1 -
Generate Rows
1 -
Getting Started
1 -
Google Analytics
140 -
grouping
1 -
Guidelines
11 -
Hello Everyone !
2 -
Help
4,113 -
How do I colour fields in a row based on a value in another column
1 -
How-To
1 -
Hub 20.4
2 -
I am new to Alteryx.
1 -
identifier
1 -
In Database
855 -
In-Database
1 -
Input
3,714 -
Input data
2 -
Inserting New Rows
1 -
Install
3 -
Installation
305 -
Interface
2 -
Interface Tools
1,646 -
Introduction
5 -
Iterative Macro
951 -
Jira connector
1 -
Join
1,738 -
knowledge base
1 -
Licenses
1 -
Licensing
210 -
List Runner
1 -
Loaders
12 -
Loaders SDK
1 -
Location Optimizer
52 -
Lookup
1 -
Machine Learning
230 -
Macro
2 -
Macros
2,501 -
Mapping
1 -
Marketo
12 -
Marketplace
4 -
matching
1 -
Merging
1 -
MongoDB
66 -
Multiple variable creation
1 -
MultiRowFormula
1 -
Need assistance
1 -
need help :How find a specific string in the all the column of excel and return that clmn
1 -
Need help on Formula Tool
1 -
network
1 -
News
1 -
None of your Business
1 -
Numeric values not appearing
1 -
ODBC
1 -
Off-Topic
14 -
Office of Finance
1 -
Oil & Gas
1 -
Optimization
647 -
Output
4,506 -
Output Data
1 -
package
1 -
Parse
2,101 -
Pattern Matching
1 -
People Person
6 -
percentiles
1 -
Power BI
197 -
practice exercises
1 -
Predictive
2 -
Predictive Analysis
820 -
Predictive Analytics
1 -
Preparation
4,633 -
Prescriptive Analytics
185 -
Publish
230 -
Publishing
2 -
Python
728 -
Qlik
36 -
quartiles
1 -
query editor
1 -
Question
18 -
Questions
1 -
R Tool
452 -
refresh issue
1 -
RegEx
2,107 -
Remove column
1 -
Reporting
2,114 -
Resource
15 -
RestAPI
1 -
Role Management
3 -
Run Command
501 -
Run Workflows
10 -
Runtime
1 -
Salesforce
244 -
Sampling
1 -
Schedule Workflows
3 -
Scheduler
372 -
Scientist
1 -
Search
3 -
Search Feedback
20 -
Server
525 -
Settings
759 -
Setup & Configuration
47 -
Sharepoint
465 -
Sharing
2 -
Sharing & Reuse
1 -
Snowflake
1 -
Spatial
1 -
Spatial Analysis
557 -
Student
9 -
Styling Issue
1 -
Subtotal
1 -
System Administration
1 -
Tableau
462 -
Tables
1 -
Technology
1 -
Text Mining
410 -
Thumbnail
1 -
Thursday Thought
10 -
Time Series
397 -
Time Series Forecasting
1 -
Tips and Tricks
3,783 -
Tool Improvement
1 -
Topic of Interest
40 -
Transformation
3,214 -
Transforming
3 -
Transpose
1 -
Truncating number from a string
1 -
Twitter
24 -
Udacity
85 -
Unique
2 -
Unsure on approach
1 -
Update
1 -
Updates
2 -
Upgrades
1 -
URL
1 -
Use Cases
1 -
User Interface
21 -
User Management
4 -
Video
2 -
VideoID
1 -
Vlookup
1 -
Weekly Challenge
1 -
Weibull Distribution Weibull.Dist
1 -
Word count
1 -
Workflow
8,474 -
Workflows
1 -
YearFrac
1 -
YouTube
1 -
YTD and QTD
1
- « Previous
- Next »