Alteryx Designer Desktop Discussions
Find answers, ask questions, and share expertise about Alteryx Designer Desktop and Intelligence Suite.- Community
- :
- Community
- :
- Participate
- :
- Discussions
- :
- Designer Desktop
- :
- Elastic Net/Lasso/Ridge Regression
Elastic Net/Lasso/Ridge Regression
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hello,
I want to run elastic net regression, which I am trying to run by using "GLMNET" package in "R Tool". I do not have much experiece in R and I am running into errors "S4 class is not subsettable" and I am sure I will get more once I solve this one.
Has any one sucessfully used Lasso/Ridge/Elasticnet regression in Alteryx? what package did you use and can you share with me how you did it ?
Thanks!
Ashish
Solved! Go to Solution.
- Labels:
-
Predictive Analysis
-
R Tool
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @bridgetT
Thanks for your help in past.
I am at this thread again as I am facing another problem in the same workwoflow. I am trying to use the weights in glmnet and I am getting error: R (2) number of elements in weights (0) not equal to the number of rows of x (744).
Pasted below is the code that you helped me with earlier.
I have left your comments in from past to help remember the context.
I have added the weight variable in the code (highlighted) and I am not sure why its not working. I have tried this code in 'Rstudio' and it works in 'Rstudio' as is.
# Read the data stream into R
the.data_test <- read.Alteryx("#1", mode="data.frame")
the.data_train <- read.Alteryx("#2", mode="data.frame")
require(glmnet)
require(stats)
#Instead of starting with the empty dataframes as you did, we'll initialize dataframes of the correct size to hold all the data.
#This data frame will hold the test data. Additionally, the last column will hold the predicted values for the test data.
#It'll play the same role as your newdf.
number.entries <- (NROW(the.data_test) * (NCOL(the.data_test) + 1))
#test.data.out <- as.data.frame(matrix(vector(mode = "numeric", length = number.entries), nrow = NROW(the.data_test), ncol = (NCOL(the.data_test) + 1)))
test.data.out <- as.data.frame(matrix(vector(mode = "numeric"), nrow = 0, ncol = (NCOL(the.data_test) + 1)))
#Create Group variable with unique group values to use to iterate through the loop
Grp <- unique(the.data_train[,34])
#Now that we've created Grp, we can initialize coef1.
#First we initialize the number of entries that will be in the coefficient dataframe.
#You seem to want the entire coefficient matrix for each iteration.
#(Recall that each call to glmnet will find your coefficients through an iterative process. The last step of this process will yield your "best" coefficients.)
#Since you didn't supply a value for the number of lambda's (ie the number of iterations), the program will choose the default number of 100.
number.coef.entries <- sum(!is.na(Grp)) * 19
###Now we can initialize the dataframe to have all 0's. On each iteration of the for loop, we will update a new row.
##all.coefs <- as.data.frame(matrix(vector(mode = "numeric", length = number.coef.entries), nrow = (sum(!is.na(Grp))), ncol = 18))
all.coefs <-as.data.frame(matrix(NA,nrow = 0, ncol = 19))
#Initialize the counter to mark where we are on updating test.data.out
current.test.row <- 1
#This one will mark which row of the coef matrix we're on
current.coef.row <- 0
for (i in Grp){
filt_data <- the.data_train[which(the.data_train$Group == i),]
if (nrow(filt_data) < 20) next()
t_pred_matrix <- as.matrix(filt_data[,16:33])
#t_pred_matrix <- as.matrix(scale(pred_matrix))
target_var <- as.matrix(filt_data[,15])
#I took the liberty of eliminating the lambda parameter here.
#The reference manual for glmnet says that typical usage is to not supply a value for lambda and to let the program compute it.
#If you do want to supply values, you need to supply a decreasing sequence of lambda values rather than a single value.
##weights1 <- vector(mode = "numeric") ## I tried to use vectors in hop that it will resolve the issue
##weights1 <- c(weights1,filt_data$weights)
fit <- glmnet(t_pred_matrix, target_var,family = 'gaussian',weights = filt_data$weights, alpha = 0.5)
filt_data_test <- the.data_test[which(the.data_test$Group == i),]
if (nrow(filt_data_test) < 2) next()
npred_matrix <- as.matrix(filt_data_test[,16:33])
pred_change <- predict(fit,newx = npred_matrix)
filt_data_test$pred_chng <- pred_change[,ncol(pred_change)]
#test.data.out[current.test.row: current.test.row +NROW(filt_data_test),] <- filt_data_test
test.data.out <- rbind(test.data.out, filt_data_test[nrow(filt_data_test),])
coef1 <- as.data.frame(t(as.matrix(coef(fit))))
coef1$name <- i
all.coefs <- rbind(all.coefs,coef1[nrow(coef1),])
# coeff.start <- current.coef.row + 1
# coeff.end <- current.coef.row * 100
# all.coefs[coeff.start:coeff.end,] <- coef1
# current.test.row <- current.test.row + NROW(filt_data_test)
# current.coef.row <- current.coef.row + 100
}
#Filter out any potential cases where the glmnet algorithm terminated before 100 iterations:
write.Alteryx(all.coefs,1)
write.Alteryx(test.data.out,2)
Looking forward for help again.
Thanks,
Ashish Singhal
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @Ashish,
Did you get that same error with both approaches you tried (making the weights1 vector, and just directly trying to take the weights column)? It seems like R thinks that you have no columns in filt_data called weights. But I'm a bit mystified as to why the code works in RStudio, but not in Alteryx. Did you do anything in your workflow before reading the data in, or did you read it directly from the same file you read from in RStudio? The only time I've seen differences between RStudio and the Alteryx R tool is when strings are treated as factors in Alteryx but not RStudio. Does your weights vector have any missing values in it that got coded as the string "NA" or some other string? Then Alteryx would probably read it in as a factor. The weights argument in glmnet() only takes numeric values, but I'd expect to see an error complaining that you tried to give it non-numeric values rather than that you gave it no values at all. But maybe it has some surprising error handling.
Best,
Bridget
Research Scientist, Analytic Products
Alteryx
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @Bridget,
Thanks for your thoughts.
Yes, I am getting the same error in both the approaches.
I am reading the same file in from the database, but there is difference in how I am adding the weight variable. In alteryx I am using the formula tool to add the weight variable, while in RStudio I am using R statements to create the weight variable.
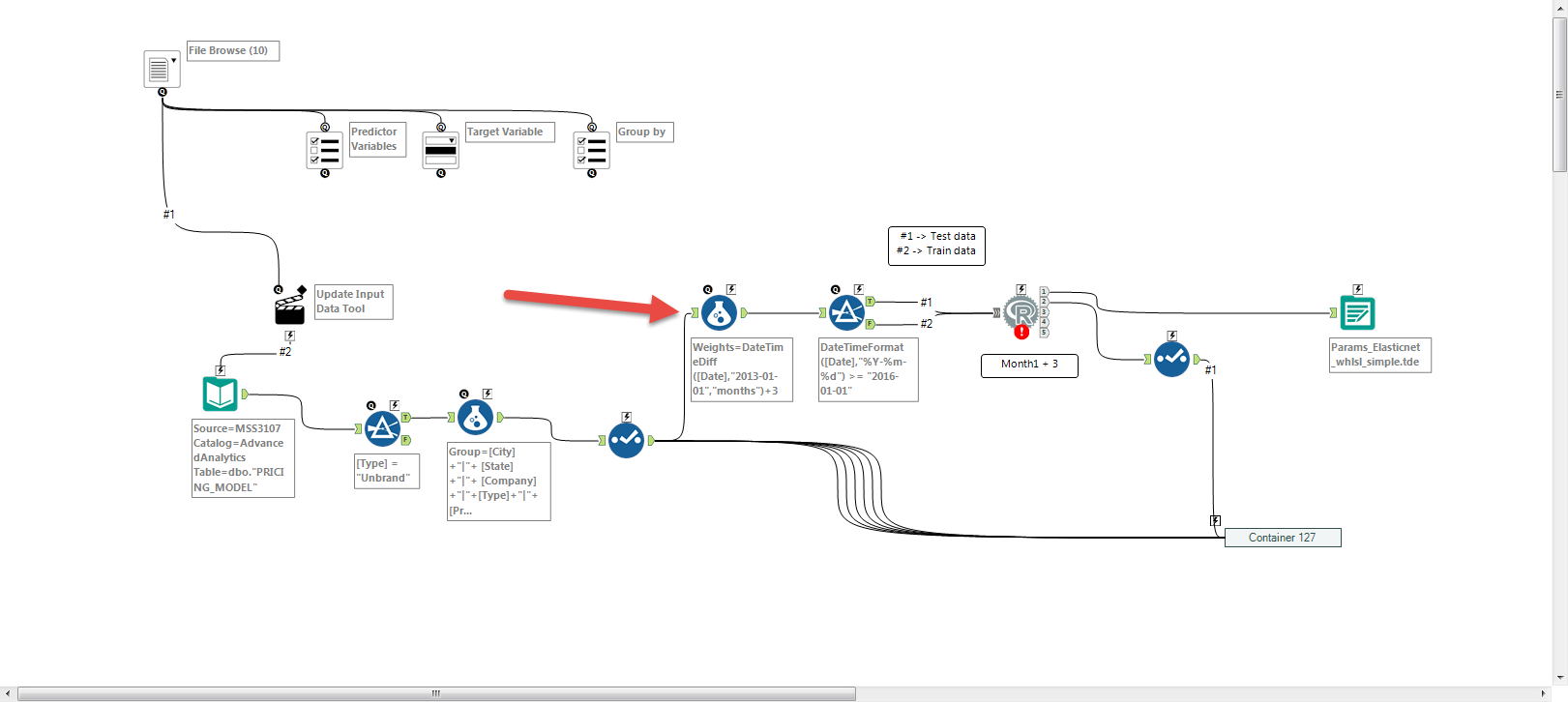
R code and Alterxy workflow image pasted below.
cn <- odbcDriverConnect(connection="Driver={SQL Server Native Client 11.0};server=MSS3107;database=AdvancedAnalytics;trusted_connection=yes;")
Input <- sqlQuery(cn, "select * from [AdvancedAnalytics].[dbo].[PRICING_MODEL]")
# Create group variable by concatinating city and company
Input$Group <- paste(Input$City,Input$State, Input$Company, Input$Type, Input$`Prediction Commodity`, sep = "|")
Input$weights <- difftime(Input$Date,"2013-01-01",units = "weeks")+20
the.data_test <- Input[ Input$Date >= as.POSIXct("2015-12-31"),]
the.data_train <- Input[ Input$Date <= as.POSIXct("2015-12-31"),]
I am doing this because I wanted to compare results of different weighting scheme's, and I have duplicated the modelling piece of the workflow with different formula for weights (hidden in container). while in Rstudio I was just trying to run it once with any weight. There are no missing values or NA in the weights variable.
Thanks,
Ashish
(P.S. Can you please tell me how to use @ to notify someone when you want to mention them)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @Ashish,
It looks like you're naming the variable Weights (with a capital W) with the Formula tool. R is case sensitive, so trying to extract the column weights when you really want the column Weights won't give you anything. You should either change the name to weights with the Formula tool or edit your R code to extract Weights.
To tag people, just type @ and then the first letter of their name. Then a list of relevant people should show up, and you can click on the person you want to tag.
Best,
Bridget
Research Scientist, Analytic Products
Alteryx
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Thanks a lot Bridget,
It resolved the issue.
I am ashamed that I spent close to 8 hours (almost whole day yesterday) trying to fix it.
Have no words to explain my facepalm right now.
Regards,
Ashish
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
You're welcome, @Ashish! And don't feel bad! Sometimes it just takes another set of eyes to notice these things. I've definitely been in similar situations myself.
Research Scientist, Analytic Products
Alteryx
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @BridgetT
In another experiment I am in a similar situation where the code works fine in R studio but there is a warning in Alteryx, which leads to undesirable results.
I am trying to subset my data based on date within R tool, the reason I am doing it in R tool (instead of using alteryx filter tool) is because I want to use function model.matrix for categorical variable before splitting my data and the reason for that is subsetted datasets may or may not have all the factors.
Pasted is the code I am trying
# Read the data stream into R
Input <- read.Alteryx("#1", mode="data.frame")
season_matrix <- model.matrix(Input$Change ~Input$Season)[,-1]
Input <- cbind(Input,season_matrix)
#Input$Date <- as.Date(Input$Date)
the.data_test <- Input[ Input$Date >= as.POSIXct("2015-12-31"),]
the.data_train <- Input[ Input$Date <= as.POSIXct("2015-12-31"),]
#the.data_test <- Input[ format(Input$Date,"%Y-%m-%d") >= "2015-12-31",]
#the.data_train <- Input[ format(Input$Date,"%Y-%m-%d") <= "2015-12-31",]
#the.data_test <- subset(Input , as.Date(Date) > as.POSIXct("2015-12-31"))
#the.data_train <- subset(Input , as.Date(Date) <= as.POSIXct("2015-12-31"))But I am getting this warning.
R (34) In `[.data.frame`(Input, Input$Date >= as.POSIXct("2015-12-31"), :
R (34) Incompatible methods ("Ops.factor", "Ops.POSIXt") for ">="
Thanks in advance for your help.
Regards,
Ashish
P.S. using @ symbol seems not to work for me to notify you :(
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
ok, I figured out the fix for the problem. Although I do not fully comprehend why it was not working earlier and why its working now, but somehow it worked.
I used as.Date function on both sides of operator to coerce the type.
require(glmnet)
require(stats)
# Read the data stream into R
Input <- read.Alteryx("#1", mode="data.frame")
season_matrix <- model.matrix(Input$Change ~Input$Season)[,-1]
Input <- cbind(Input,season_matrix)
#Input$Date <- as.Date(Input$Date)
#the.data_test <- Input[ Input$Date >= as.POSIXct("2015-12-31"),]
#the.data_train <- Input[ Input$Date <= as.POSIXct("2015-12-31"),]
the.data_test <- Input[ as.Date(Input$Date) >= as.Date("2015-12-31"),]
the.data_train <- Input[ as.Date(Input$Date) <= as.Date("2015-12-31"),]The "Date" variable in the dataset was already of type date so I did not suspect it earlier.
Also when I tried formula tool create a new Date variable date format that did not work either.
I am relieved that I've found a fix but I lack the understanding what works and why.
Thanks,
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi @Ashish,
The problem is that the read.Alteryx function does not natively read in date types in Alteryx as date types in R. I'm attaching a small example to show this phenomenon. (Note that you'll need to have the package DescTools installed in order to run it.) I'm not sure exactly how you got the date field to read in as a date type on RStudio but not the R tool, though. What type of file were you using?
Best,
Bridget
Research Scientist, Analytic Products
Alteryx
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Hi Bridget,
Thanks for the example you shared. I also tried this workflow on my file and it converty the date into a V_Wstring
My source file is a SQL Sever table and the Date filed is of the format " yyyy-mm-dd hh:mm:ss",
I use following connection string to connect and it recognize the filed as date time.
cn <- odbcDriverConnect(connection="Driver={SQL Server Native Client 11.0};server=MSS1234;database=AdvancedAnalytics;trusted_connection=yes;")
Input <- sqlQuery(cn, "select * from [AdvancedAnalytics].[dbo].[PRICING_MODEL]")
Regards,
Ashish
-
AAH
1 -
AAH Welcome
2 -
Academy
24 -
ADAPT
82 -
Add column
1 -
Administration
20 -
Adobe
176 -
Advanced Analytics
1 -
Advent of Code
5 -
Alias Manager
70 -
Alteryx
1 -
Alteryx 2020.1
3 -
Alteryx Academy
3 -
Alteryx Analytics
1 -
Alteryx Analytics Hub
2 -
Alteryx Community Introduction - MSA student at CSUF
1 -
Alteryx Connect
1 -
Alteryx Designer
44 -
Alteryx Engine
1 -
Alteryx Gallery
1 -
Alteryx Hub
1 -
alteryx open source
1 -
Alteryx Post response
1 -
Alteryx Practice
134 -
Alteryx team
1 -
Alteryx Tools
1 -
AlteryxForGood
1 -
Amazon s3
138 -
AMP Engine
191 -
ANALYSTE INNOVATEUR
1 -
Analytic App Support
1 -
Analytic Apps
17 -
Analytic Apps ACT
1 -
Analytics
2 -
Analyzer
17 -
Announcement
4 -
API
1,038 -
App
1 -
App Builder
43 -
Append Fields
1 -
Apps
1,167 -
Archiving process
1 -
ARIMA
1 -
Assigning metadata to CSV
1 -
Authentication
4 -
Automatic Update
1 -
Automating
3 -
Banking
1 -
Base64Encoding
1 -
Basic Table Reporting
1 -
Batch Macro
1,271 -
Beginner
1 -
Behavior Analysis
217 -
Best Practices
2,413 -
BI + Analytics + Data Science
1 -
Book Worm
2 -
Bug
622 -
Bugs & Issues
2 -
Calgary
59 -
CASS
46 -
Cat Person
1 -
Category Documentation
1 -
Category Input Output
2 -
Certification
4 -
Chained App
235 -
Challenge
7 -
Charting
1 -
Clients
3 -
Clustering
1 -
Common Use Cases
3,388 -
Communications
1 -
Community
188 -
Computer Vision
45 -
Concatenate
1 -
Conditional Column
1 -
Conditional statement
1 -
CONNECT AND SOLVE
1 -
Connecting
6 -
Connectors
1,180 -
Content Management
8 -
Contest
6 -
Conversation Starter
17 -
copy
1 -
COVID-19
4 -
Create a new spreadsheet by using exising data set
1 -
Credential Management
3 -
Curious*Little
1 -
Custom Formula Function
1 -
Custom Tools
1,720 -
Dash Board Creation
1 -
Data Analyse
1 -
Data Analysis
2 -
Data Analytics
1 -
Data Challenge
83 -
Data Cleansing
4 -
Data Connection
1 -
Data Investigation
3,061 -
Data Load
1 -
Data Science
38 -
Database Connection
1,898 -
Database Connections
5 -
Datasets
4,577 -
Date
3 -
Date and Time
3 -
date format
2 -
Date selection
2 -
Date Time
2,880 -
Dateformat
1 -
dates
1 -
datetimeparse
2 -
Defect
2 -
Demographic Analysis
173 -
Designer
1 -
Designer Cloud
473 -
Designer Integration
60 -
Developer
3,646 -
Developer Tools
2,919 -
Discussion
2 -
Documentation
453 -
Dog Person
4 -
Download
906 -
Duplicates rows
1 -
Duplicating rows
1 -
Dynamic
1 -
Dynamic Input
1 -
Dynamic Name
1 -
Dynamic Processing
2,539 -
dynamic replace
1 -
dynamically create tables for input files
1 -
Dynamically select column from excel
1 -
Email
743 -
Email Notification
1 -
Email Tool
2 -
Embed
1 -
embedded
1 -
Engine
129 -
Enhancement
3 -
Enhancements
2 -
Error Message
1,977 -
Error Messages
6 -
ETS
1 -
Events
178 -
Excel
1 -
Excel dynamically merge
1 -
Excel Macro
1 -
Excel Users
1 -
Explorer
2 -
Expression
1,695 -
extract data
1 -
Feature Request
1 -
Filter
1 -
filter join
1 -
Financial Services
1 -
Foodie
2 -
Formula
2 -
formula or filter
1 -
Formula Tool
4 -
Formulas
2 -
Fun
4 -
Fuzzy Match
614 -
Fuzzy Matching
1 -
Gallery
590 -
General
93 -
General Suggestion
1 -
Generate Row and Multi-Row Formulas
1 -
Generate Rows
1 -
Getting Started
1 -
Google Analytics
140 -
grouping
1 -
Guidelines
11 -
Hello Everyone !
2 -
Help
4,112 -
How do I colour fields in a row based on a value in another column
1 -
How-To
1 -
Hub 20.4
2 -
I am new to Alteryx.
1 -
identifier
1 -
In Database
854 -
In-Database
1 -
Input
3,713 -
Input data
2 -
Inserting New Rows
1 -
Install
3 -
Installation
305 -
Interface
2 -
Interface Tools
1,645 -
Introduction
5 -
Iterative Macro
951 -
Jira connector
1 -
Join
1,737 -
knowledge base
1 -
Licenses
1 -
Licensing
210 -
List Runner
1 -
Loaders
12 -
Loaders SDK
1 -
Location Optimizer
52 -
Lookup
1 -
Machine Learning
230 -
Macro
2 -
Macros
2,500 -
Mapping
1 -
Marketo
12 -
Marketplace
4 -
matching
1 -
Merging
1 -
MongoDB
66 -
Multiple variable creation
1 -
MultiRowFormula
1 -
Need assistance
1 -
need help :How find a specific string in the all the column of excel and return that clmn
1 -
Need help on Formula Tool
1 -
network
1 -
News
1 -
None of your Business
1 -
Numeric values not appearing
1 -
ODBC
1 -
Off-Topic
14 -
Office of Finance
1 -
Oil & Gas
1 -
Optimization
647 -
Output
4,504 -
Output Data
1 -
package
1 -
Parse
2,101 -
Pattern Matching
1 -
People Person
6 -
percentiles
1 -
Power BI
197 -
practice exercises
1 -
Predictive
2 -
Predictive Analysis
820 -
Predictive Analytics
1 -
Preparation
4,632 -
Prescriptive Analytics
185 -
Publish
230 -
Publishing
2 -
Python
728 -
Qlik
36 -
quartiles
1 -
query editor
1 -
Question
18 -
Questions
1 -
R Tool
452 -
refresh issue
1 -
RegEx
2,106 -
Remove column
1 -
Reporting
2,113 -
Resource
15 -
RestAPI
1 -
Role Management
3 -
Run Command
501 -
Run Workflows
10 -
Runtime
1 -
Salesforce
243 -
Sampling
1 -
Schedule Workflows
3 -
Scheduler
372 -
Scientist
1 -
Search
3 -
Search Feedback
20 -
Server
524 -
Settings
759 -
Setup & Configuration
47 -
Sharepoint
465 -
Sharing
2 -
Sharing & Reuse
1 -
Snowflake
1 -
Spatial
1 -
Spatial Analysis
557 -
Student
9 -
Styling Issue
1 -
Subtotal
1 -
System Administration
1 -
Tableau
462 -
Tables
1 -
Technology
1 -
Text Mining
410 -
Thumbnail
1 -
Thursday Thought
10 -
Time Series
397 -
Time Series Forecasting
1 -
Tips and Tricks
3,782 -
Tool Improvement
1 -
Topic of Interest
40 -
Transformation
3,214 -
Transforming
3 -
Transpose
1 -
Truncating number from a string
1 -
Twitter
24 -
Udacity
85 -
Unique
2 -
Unsure on approach
1 -
Update
1 -
Updates
2 -
Upgrades
1 -
URL
1 -
Use Cases
1 -
User Interface
21 -
User Management
4 -
Video
2 -
VideoID
1 -
Vlookup
1 -
Weekly Challenge
1 -
Weibull Distribution Weibull.Dist
1 -
Word count
1 -
Workflow
8,473 -
Workflows
1 -
YearFrac
1 -
YouTube
1 -
YTD and QTD
1
- « Previous
- Next »